Overexpression of KAI1 Protein in Diabetic Skin Tissues
Article information
Abstract
Background
Patients with diabetes mellitus often have a difficult life, suffering from foot ulceration or amputation. Diabetes is characterized by chronic inflammation, and one of the features of inflammation is hypoxia. Recently, it has been reported that KAI1 is a hypoxia target gene. There is no published research on hypoxia-related KAI1 protein levels in human diabetic skin. Therefore, we have investigated the expression of KAI1 protein in diabetic skin tissue in vivo.
Methods
The expression of KAI1 protein was evaluated by western blotting in 6 diabetic skin tissue samples and 6 normal skin samples. Immunohistochemical staining was carried out to identify KAI1 expression.
Results
The western blotting revealed significantly increased expression of the KAI1 protein in diabetic skin tissues as compared to normal skin tissues. Immunohistochemical examination demonstrated that KAI1 was expressed in all diabetic skin tissues with moderate-to-strong positivity and weakly expressed in normal skin tissues.
Conclusions
Our data suggest that a high expression of the KAI1 protein can be observed in diabetic skin tissue. To the best of our knowledge, this is the first report suggesting that KAI1 protein expression in diabetic skin tissues may be associated with chronic inflammatory states and hypoxia.
INTRODUCTION
Diabetes mellitus causes significant health problems. It has been reported that the oxygen concentration in pancreatic β-cells controls the activity of the pancreatic cells through hypoxia-inducible factor (HIF)-α [1,2]. Tissue hypoxia produces poor glucose and insulin regulation, and insulin resistance, which can lead to diabetes. Hypoxia also has many other detrimental effects, such as oxidative stress, free radicals, and chronic inflammation. Ahmad et al. [3] reported that diabetes is characterized by a chronic inflammatory state. Hypoxia is a characteristic feature of inflammation [4,5]. Thus, the chronic inflammation of diabetes makes hypoxia a key process [6]. A hypoxic environment triggers cells to up-regulate HIF-α subunits [7]. Cells are exposed to abnormal molecular levels of of HIF-α, which results in dysregulated signal transduction.
KAI1 was first found in a T-cell activation study [8]. The expression product of KAI1 is a glycoprotein, which is distributed in the surface of most cells in the body. The KAI1 gene is located on chromosome 11p11.2 and is composed of 10 exons and 9 introns [9]. According to Kim et al. [10], a promoter of the KAI1 gene contains a functional hypoxia-response element (HRE). They suggest that KAI1 is a hypoxia target gene and that hypoxia induces KAI1 expression. Hypoxia target genes containing functional HRE influence cell signaling including glucose metabolism [11]. It is known that diabetic complications are related to hypoxic conditions. However, little is known about the role of KAI1 in diabetic skin in vivo. In light of this, we explored the expression of the KAI1 protein in order to determine whether it is down-regulated or up-regulated in normal and diabetic skin tissues.
METHODS
Tissue samples
The Institutional Review Board of Soonchunhyang University Hospital in Korea reviewed and approved this research protocol involving the use of tissue samples. A total of 6 normal skin tissue samples and 6 diabetic skin samples were obtained from patients who underwent surgery between December 2012 and June 2013 in the Departments of Plastic and Reconstructive Surgery at Soonchunhyang University Hospital. Informed consent was obtained from the patients before surgery. The normal skin tissue samples were collected from the lower leg of patients who had undergone reconstruction with a flap. Diabetic skin samples were obtained from patients undergoing amputation surgery. A portion of the specimens were frozen in liquid nitrogen immediately after resection, and stored at -80℃. For this study, the stored formalin-fixed, paraffin-embedded samples were used.
Immunohistochemical staining
Paraffin sections were deparaffinized in xylene, rehydrated in 10 mM citrate buffer (pH 6.0), and heated in a microwave oven for 15 minutes to restore the antigens. To suppress endogenous peroxidase within the tissues, the samples were treated with 3% peroxide for 5 min and then with a blocking solution for 30 min. Slides were incubated with the primary antibody in a humidity chamber for 60 min. Tissue staining was visualized with 3,3'-diaminobenzidine (ScyTek, Logan, UT, USA) substrate chromogen solution. Primary mouse KAI1 (G-2) antibody (catalog number SC-17752) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Assessment of immunohistochemical staining
The reactivity was measured for estimating the intensity of labeling (-, 1+, 2+, and 3+) and the percentage of immunohistochemically stained cells (<25%, 25-75%, and >75%). The assessment was decided by two sets of independent investigators.
Western blot analysis
Tissue samples were homogenized in a WCE buffer (25 mM HEPES [pH 7.7], 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM ethylenediaminetetraacetic acid, 0.1% Triton X-100, 0.5 mM dithiothreitol, 20 mM glycerophosphate, 0.1 mM Na3VO4, 2 g/mL leupeptin, 2 g/mL aprotinin, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail tablet [Boehringer Mannheim]). The tissue suspension was rotated at 4℃ for 10 min. The supernatants were collected, kept at -70℃, and used for western blotting. The proteins from the tissue were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using NuPAGE 4-12% bis-Tris gels (NP0335Box, Invitrogen, Seoul, Korea) and then transferred to an Immobilon-P membrane (Immobilon-P, Millipore Korea Co. Ltd., Seoul, Korea). The membrane was blocked using 5% bovine serum albumin in TBST (20 mM Tris, pH 7.6, 130 mM NaCl, and 0.1% Tween 20) solution. It was reacted with the primary mouse KAI1 (G-2) antibody (catalog number SC-17752, Santa Cruz Biotechnology) diluted to a 1:1,000 concentration, stored at 4℃ for 16 hours, and mixed with a washing buffer and a TBST buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.05% Tween 20), and then washed well 4 times: for 10 minutes, 10 minutes, 15 minutes, and 15 minutes. It was then reacted for 1 hour with a 1:10,000 dilution of the secondary antibody (species-specific whole anti-mouse IgG conjugated to horseradish peroxidase; SC-2005; Santa Cruz Biotechnology). After the reaction with the secondary antibody, it was again washed well 4 times: for 10 minutes, 10 minutes, 15 minutes, and 15 minutes. Proteins on the membrane were detected using the enhanced chemiluminescence solution kit (Amersham Pharmacia Biotech, Amersham, Buckinghamshire, UK). The membranes were stripped and reblotted with an anti-actin antibody (Sigma, St. Louis, MI, USA; Catalog number A5441).
Assessment of western blot analysis
The relative abundance of each protein expression was analyzed by Phosphor-Imager software (TINA, Raytest, Straubenhardt, Germany). The measured amounts of the expression of diabetic skin and normal skin tissues were compared.
Statistical analysis
The data from the Raytest TINA software were analyzed using SPSS ver. 17.0 (SPSS Inc., Chicago, IL, USA), and statistical significance was set at a P-value of less than 0.05. For a comparison of the non-normally distributed variables, we used the Mann-Whitney U test.
RESULTS
Immunohistochemical examination
The immunohistochemical study showed that the KAI1 protein was more markedly expressed in diabetic skin with moderate-to-strong positivity (Table 1). All 6 cases of diabetic skin tissues displayed 2+ to 3+ staining intensity for KAI1. Of the 6 labeled cases, 0 (0%) had fewer than 25% of the cells stained, 3 cases (50%) had 25% to 75% of the cells stained, and 3 cases (50%) had more than 75% of the cells stained. In contrast with the diabetic skin tissues, KAI1 was weakly expressed in the normal skin tissue. Four of the 6 cases of normal skin tissue displayed 1+ staining intensity for KAI1. The 4 cases had fewer than 25% of the cells stained.
In the diabetic skin tissue samples, KAI1 immunoreactivity was present in the peripheral membrane staining of keratinocytes at the epidermis and in appendageal structures (Fig. 1).
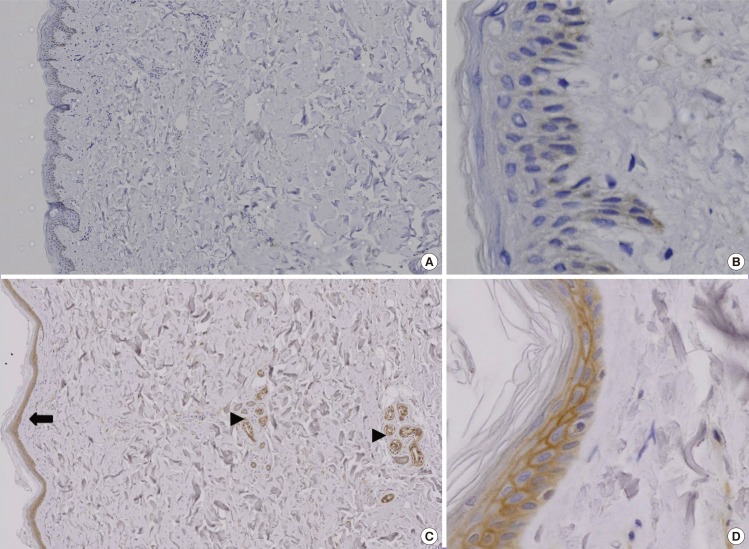
Immunohistochemical study of KAI1 protein
The color brown denotes a positive stain. (A) Fewer than 25% of the cells in the normal skin tissue were stained (immunochemical stain, 20×). (B) KAI1 was weakly stained in the normal skin tissue (immunochemical stain, 200×). (C) The diabetic skin tissue displayed increased expression of KAI1. KAI1 immunoreactivity was present in the peripheral membranous staining of keratinocytes (black arrow heads) and appendageal structures (black arrow) (immunochemical stain, 20×). (D) In the diabetic skin tissue samples, KAI1 immunoreactivity was present in the peripheral membrane staining of keratinocytes (immunochemical stain, 200×).
Western blot analysis
Western blot analysis showed that the KAI1 protein was more expressed in diabetic skin tissues compared with normal human skin tissues (Fig. 2). The expression amount score in western blotting was analyzed by the Mann-Whitney U test, and the median of KAI1 was 0.08 (range, 0.07-0.09) in normal skin and 0.60 (range, 0.29-0.74) in human diabetic skin tissue (Fig. 3). There were significant differences in KAI1 protein expression between the normal skin and diabetic skin (P<0.05).
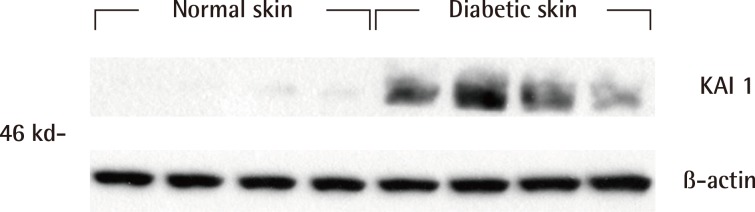
Western blot analysis of KAI1 protein
Western blot analysis showed that KAI1 was more markedly expressed in the diabetic skin tissues than in the normal human skin tissues.
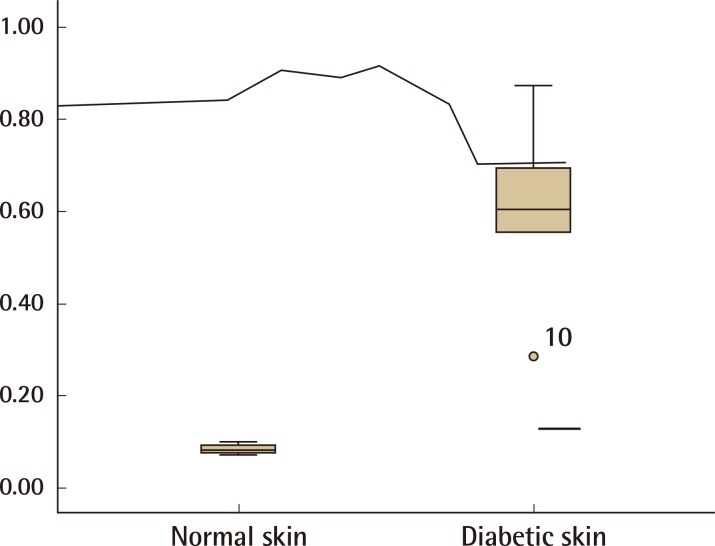
The score of relative KAI1 protein expression
The relative protein expression of KAI1 was analyzed with the Mann-Whitney U test, and the median of KAI1 was found to be 0.08 (range, 0.07-0.09) in normal skin and 0.60 (range, 0.29-0.74) in human diabetic skin tissue. There were significant differences in the KAI1 protein expression between normal skin and diabetic skin (P<0.05).
DISCUSSION
KAI1, also called CD82, C33, CD82, and ST6, is a widely expressed plasma membrane glycoprotein and belongs to the structurally distinct transmembrane 4 superfamily (TM4SF). TM4SF members form one small loop and one large extracellular loop with residues that are prone to N-linked glycosylation or post-translational phosphorylation. Due to the glycosylation, the KAI1 protein has a molecular weight of 46-60 kDa [12]. The mRNA levels of KAI1 are highly expressed in the spleen, lung, liver, placenta, prostate, and kidney, and moderately in the skeletal muscle, thymus, and pancreas; its expression is low in the brain, heart, uterus, stomach, and ovary [13,14]. However, little is known about the expression of KAI1 in skin tissues.
Diabetes induces chronic hypoxia and an excessive inflammatory response [15]. At the cellular level, hypoxic microenvironments affect various pathophysiological conditions including inflammation, embryogenesis, wound healing, tumorigenesis, ischemic disease, and atherosclerosis [11,16]. Interestingly, Kim et al. [10] reported that the KAI1 gene promoter contains a functional HRE and that KAI1 protein synthesis is markedly induced in ischemic heart tissues in vivo. They suggest that KAI1 is a hypoxia target gene and hypoxia-induced KAI1 regulates a subset of hypoxia target genes. Hypoxia induces KAI1 expression by HIF-1α binding on the promoter of KAI1. HIF is a transcription factor, consisting of HIF-1α, HIF-1β, and HIF-2α subunits. Under normal conditions, HIF-α is controlled by prolyl hydroxylase, which is activated, and the hydroxylated HIF-α attaches to von Hippel-Lindau E3 ligase and undergoes proteasomal degradation [17]. However, under hypoxic states, the activity of prolyl hydroxylase is decreased and HIF-α translocates into the nucleus, binding to HIF-β [18]. The HIF-α/β complex binds to the HRE. Hypoxia target genes containing functional HRE affect diverse cellular processes including glucose metabolism, apoptosis, erythropoiesis, and transcriptional regulation [11]. Hypoxia-dependent induction of KAI1 was directly mediated by HIF-1α binding on the promoter, which subsequently caused the increased recruitment of RNA polymerase II for transcriptional activation. Kim et al. [10] reported that the failure of HIF-1α recruitment to the KAI1 promoter was observed in HIF-1α knockout mouse embryonic fibroblasts.
The inflammatory status induces the release of several factors, like interleukin (IL)-1, IL-6, tumor necrosis factor-alpha (TNF-α), and transforming growth factor-β [11]. Qian et al. [19] reported that HIF-1α was activated by IL-1β and TNF-α. Further, Kutlu et al. [20] reported a time--course microarray analysis of cytokine-induced genes in insulin-producing INS-1E cells that were exposed for 1, 2, 4, 8, 12, or 24 hours to IL-1β and interferon-γ. KAI1 (National Center for Biotechnology Information accession number AI231213) was increased eightfold at 12 hours (This finding is provided in an online Data Supplement available at http://diabetes.diabetesjournals.org/content/52/11/2701/suppl/DC1).
To the best of our knowledge, the KAI1 expression in diabetic skin tissues has not been studied previously. In fact, due to mRNA instability and post-translational modifications, alterations at the mRNA level only poorly reflect changes at the protein level. Thus, in this study, we evaluated the expression of the KAI1 protein in diabetic skin tissues using western blot analysis. The results revealed that the KAI1 protein was hardly expressed in normal skin tissues. In diabetic skin tissues, the expression of the KAI1 protein was found to be increased. Our study suggests that diabetic skin tissue induces KAI1 expression and supports the role of KAI1 under hypoxic conditions and inflammatory states. We concluded that KAI1 protein expression in diabetic skin tissues may be associated with the chronic inflammatory state and concomitant hypoxia. The exact mechanisms are still unknown. Further studies are required to elucidate the roles of KAI1 and to clarify the pathways by which these mechanisms act.
Notes
This work was supported by the Soonchunhyang University Research Fund.
No potential conflict of interest relevant to this article was reported.
